6. Agents#
This notebook aims to demonstrate how to construct LLM agents and explain their functioning. In this practical application, we will focus on extracting chemical reactions from an image.
import matextract # noqa: F401
import time
import requests
import base64
import torch
import pubchempy as pcp
from rxnscribe import RxnScribe
from rdkit import Chem
from huggingface_hub import hf_hub_download
from langchain import hub
from langchain.pydantic_v1 import BaseModel, Field
from langchain.agents import AgentExecutor
from langchain.tools import StructuredTool
from langchain.agents.react.agent import create_react_agent
from langchain_openai import ChatOpenAI
from litellm import completion
For this small demonstration we will use OpenAI newest model: GPT-4o.
model = "gpt-4o"
The GPT-4o model by OpenAI, like its predecessor, GPT-4 Turbo, is a multimodal model, meaning it can work with multimodal inputs such as text and images. Although these models work quite well for some tasks, they can not perform well when the data is field-specific. To demonstrate this, we are going to provide an image describing a chemical reaction and ask the model to extract the information describing the reaction.
To do the test, we will work with an image extracted from a work by Deem et al. [2022].
image_file = "image.png"
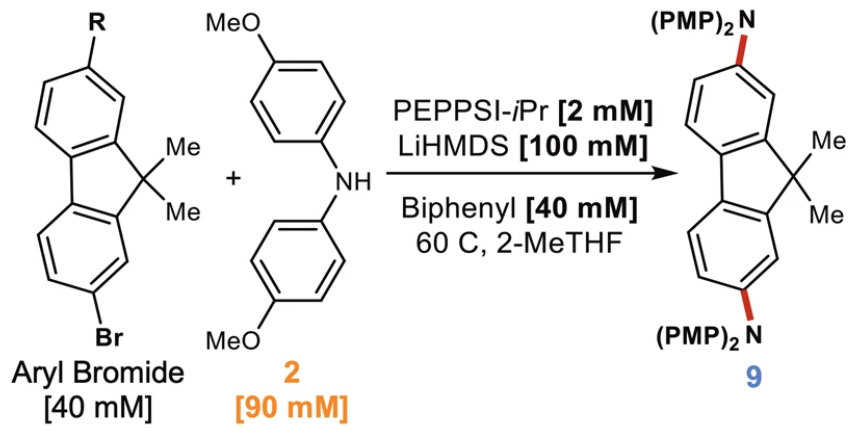
Fig. 6.1 Figure taken and cropped from Deem et al. [2022]. It was cropped to contain only the reaction itself, without all the other information that the original figure contains.#
For the record we know:
Reactant 1: 2-bromo-9,9-dimethyl-9H-fluorene with an R-chain bonded to the carbon 7
Reactant 2: 4,4′-Dimethoxydiphenylamine
Catalysts:
PEPPSI-IPr catalyst: commercial Pd catalyst
Lithium bis(trimethylsilyl)amide (LiHMDS): is primarily used as a strong non-nucleophilic base and as a ligand
Byphenyl
Solvent: 2-methyltetrahydrofuran
Product: 2,7-N,N-di(PMP)amino-9,9-dimethylfluoren or 2,7-N,N-dipolymethylpenteneamino-9,9-dimethylfluoren
To pass the image to the model through the prompt, the image needs to be encoded. To do that we use the function that OpenAI propose in their vision guidelines.
# Function to encode the image
def encode_image(image_path):
with open(image_path, "rb") as image_file:
return base64.b64encode(image_file.read()).decode("utf-8")
# Getting the base64 string
base64_image = encode_image(image_file)
After that, we generate the prompt using the function just defined to encode the image.
messages = [
{
"role": "system",
"content": "You are a chemistry expert assistant, and your task is to extract information about chemical reactions from images.",
},
{
"role": "user",
"content": [
{
"type": "text",
"text": "Please extract all the information from the next image containing a chemical reaction. For the reactants that you find, give the name or some molecular representation, such as SMILES.",
},
{
"type": "image_url",
"image_url": {
"url": f"data:image/jpeg;base64,{base64_image}",
},
},
],
},
]
And we do the completion using the OpenAI API.
response = completion(
model=model,
messages=messages,
)
response.choices[0].message.content
'The given image illustrates a chemical reaction involving an aryl bromide and another reactant (labeled 2) under specific conditions to produce a product (labeled 9). Here is the detailed information extracted from the image:\n\n### Reactants:\n1. **Aryl Bromide (40 mM)**:\n - Structure: Contains two benzene rings fused together with one of the benzene rings bonded to a bromine (Br) atom. There is a methyl (Me) group attached to each benzene ring and an R group (possibly a variable substituent) on one of the benzene rings.\n \n2. **Reactant 2 (90 mM)**:\n - Structure: Contains a biphenyl system with each phenyl ring having a methoxy group (MeO) attached. One of the phenyl rings is connected to an NH group.\n\n### Catalysts/Reagents:\n1. **PEPPSI-iPr (2 mM)** - a common ligand for facilitating palladium-catalyzed reactions.\n2. **LiHMDS (100 mM)** - Lithium hexamethyldisilazide, a strong non-nucleophilic base.\n\n### Solvent and Conditions:\n1. **Solvent**: 2-Methyltetrahydrofuran (2-MeTHF)\n2. **Temperature**: 60°C\n3. **Biphenyl (40 mM)**: Used perhaps as an additional reagent or co-catalyst component.\n\n### Product:\n- Labelled as compound 9.\n- Structure: Contains a biphenyl core similar to the reactant but with additional groups—two (PMP)₂N groups attached to the biphenyl system.\n\n### Molecular Representations:\n1. **Aryl Bromide**:\n - (SMILES): [c1cc2cc(c1)c(c(c2)C)Br]\n2. **Reactant 2**:\n - (SMILES): COc1ccc(cc1)Nc2cc(OC)ccc2 \n3. **Product 9**:\n - (SMILES, approximate): Perhaps a complex structure involving: c(biphenyl core with two (PMP)2N groups attached)\n\nThe reaction involves the conversion of an aryl bromide with a secondary amine biphenyl derivative in the presence of a palladium catalyst (PEPPSI-iPr) and a strong base (LiHMDS) under specific conditions to produce the target product (compound 9).'
The model can not give the proper name or molecular representation to the molecules. However, it can identify some functional groups and substituents of the molecules. In addition, it can surprisingly identify the solvent and all the catalysts involved in the reaction.
But luckily, some tools were developed to extract accurately this information from the images.
One of these tools is RxnScribe, which Qian et al. [2023] developed. This tool can extract chemical reactions from images. So, we will use it as a tool given to an agent that we create below. With this and other tools, we will try to use the model to extract information such as the IUPAC name and the InChI representation for the reactants involved in the reaction.
# Define a class to describe the input to the tool
class ExtractionInput(BaseModel):
image_path: str = Field(
description="Path to the image-file that contain the reaction"
)
# Define the function that will do the reaction extraction.
def extractor(image_path: str) -> list:
ckpt_path = hf_hub_download("yujieq/RxnScribe", "pix2seq_reaction_full.ckpt")
model = RxnScribe(ckpt_path, device=torch.device("cuda"))
results = model.predict_image_file(image_path, molscribe=True, ocr=True)
# Clean the output to reduce the number of tokens
for result in results:
for key, value in result.items():
for v in value:
if "molfile" in v:
v.pop("molfile")
return results
# Describe the tool for the model
image_extractor = StructuredTool.from_function(
func=extractor,
name="Reaction extractor",
description="Extract chemical reactions information such as reactants, products and catalysts from images",
args_schema=ExtractionInput,
)
If we print the variable containing the tool, we will be able to see what is going to be passed to the agent.
Reaction extractor
Extract chemical reactions information such as reactants, products and catalysts from images
{'image_path': {'title': 'Image Path', 'description': 'Path to the image-file that contain the reaction', 'type': 'string'}}
False
Similarly, we can define other tools to help the agent convert between SMILES, InChI, and the IUPAC name.
class ChemicalRepresentation(BaseModel):
smiles: str = Field(description="SMILES representation for the molecule")
def smiles_to_inchi(smiles: str) -> str:
molecule = Chem.MolFromSmiles(smiles)
return Chem.MolToInchi(molecule)
CACTUS = "https://cactus.nci.nih.gov/chemical/structure/{0}/{1}"
def smiles_to_iupac(smiles: str) -> str:
"""
Use the chemical name resolver https://cactus.nci.nih.gov/chemical/structure.
If this does not work, use pubchem.
"""
try:
time.sleep(0.001)
rep = "iupac_name"
url = CACTUS.format(smiles, rep)
response = requests.get(url, allow_redirects=True, timeout=10)
response.raise_for_status()
name = response.text
if "html" in name:
return None
return name
except Exception:
try:
compound = pcp.get_compounds(smiles, "smiles")
return compound[0].iupac_name
except Exception:
return None
smiles_to_inchi_converter = StructuredTool.from_function(
func=smiles_to_inchi,
name="Smiles to InChI",
description="Return the InChI representation of the given SMILES representation",
args_schema=ChemicalRepresentation,
)
smiles_to_iupac_converter = StructuredTool.from_function(
func=smiles_to_iupac,
name="Smiles to IUPAC",
description="Return the IUPAC name of the given SMILES representation",
args_schema=ChemicalRepresentation,
)
Once we have defined all the tools, we create a list that will be passed to the agent indicating which tools it has available.
tools = [image_extractor, smiles_to_inchi_converter, smiles_to_iupac_converter]
After creating and defining the tools, we can start constructing the agent itself by specifying the model to use. For the agent, we will use the GPT-4o model.
llm = ChatOpenAI(
temperature=0,
model_name="gpt-4o",
request_timeout=1000,
streaming="False",
)
Then we define the prompt, the agent and what it is called by LangChain as the “chain”.
# Import the ReAct prompt from the hub.
prompt = hub.pull("hwchase17/react")
# Construct the ReAct agent
agent = create_react_agent(llm, tools, prompt)
# Define the chain for the agent.
agent_executor = AgentExecutor(
agent=agent,
tools=tools,
verbose=True,
)
Notes about ReAct
Answer the following questions as best you can. You have access to the following tools:
{tools}
Use the following format:
Question: the input question you must answer
Thought: you should always think about what to do
Action: the action to take, should be one of [{tool_names}]
Action Input: the input to the action
Observation: the result of the action…(this Thought/Action/Action Input/Observation can repeat N times)
Thought: I now know the final answer
Final Answer: the final answer to the original input question\
Begin!
Question: {input}
Thought:{agent_scratchpad}
What are the variables referred in this prompt?\
toolsis a list with all the tools available. The list includes thefunc,name,descriptionandargs_schemaarguments detailed when the tools were created.\tool_namesis a list with the names of the tools available.\inputis the query from the user.\agent_scratchpadcontains the previous iterations by the agent.\
The last step before calling the agent is to define the query that we want the agent to solve.
query = f"I want the IUPAC name, the SMILES and the InChI representations for the reactants of the reaction contained in the image: {image_file}"
And finally we run the agent.
agent_executor.invoke({"input": query})
> Entering new AgentExecutor chain...
To answer this question, I need to extract the chemical reactions information from the provided image first. This will give me the reactants, products, and catalysts involved in the reaction. Once I have the reactants, I can then convert their SMILES representations to IUPAC names and InChI representations.
Action: Reaction extractor
Action Input: image.png[{'reactants': [{'category': '[Mol]', 'bbox': (0.03401700850425213, 0.01468796072044833, 0.24212106053026514, 0.8127338265314743), 'category_id': 1, 'smiles': '*c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2'}, {'category': '[Mol]', 'bbox': (0.23311655827913957, 0.011750368576358663, 0.4432216108054027, 0.8225258003451065), 'category_id': 1, 'smiles': '*.*.COc1ccc(Nc2ccc(OC)cc2)cc1'}], 'conditions': [{'category': '[Txt]', 'bbox': (0.4547273636818409, 0.20269385794218694, 0.7748874437218609, 0.3897205577825623), 'category_id': 2, 'text': ['PEPPSI-iPr [2 mM]', 'LiHMDS [100 mM]']}, {'category': '[Txt]', 'bbox': (0.4627313656828414, 0.42986765041845443, 0.76088044022011, 0.6022063895383815), 'category_id': 2, 'text': ['Biphenyl [40 mM]', '60 C, 2-MeTHF']}], 'products': [{'category': '[Mol]', 'bbox': (0.7563781890945472, 0.006854381669542554, 1.0, 0.8176298134382903), 'category_id': 1, 'smiles': 'CC1(C)c2cc(N)ccc2-c2ccc(N)cc21.I.II'}]}]The extracted information from the image provides the SMILES representations of the reactants. Here are the reactants:
1. `*c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2`
2. `*.*.COc1ccc(Nc2ccc(OC)cc2)cc1`
Next, I will convert these SMILES representations to their IUPAC names and InChI representations.
Let's start with the first reactant.
Action: Smiles to IUPAC
Action Input: *c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2NoneIt seems there was an issue with the conversion of the first SMILES representation to its IUPAC name. I will try converting the second reactant's SMILES representation to its IUPAC name.
Action: Smiles to IUPAC
Action Input: *.*.COc1ccc(Nc2ccc(OC)cc2)cc1NoneIt appears that the SMILES representations extracted from the image might contain some non-standard characters or formatting issues that are causing the conversion tools to fail. I will manually inspect and clean the SMILES strings before attempting the conversion again.
The first reactant's SMILES string is `*c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2`. The asterisk `*` might be a placeholder or an error. I will remove it and try converting the cleaned SMILES string `c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2`.
The second reactant's SMILES string is `*.*.COc1ccc(Nc2ccc(OC)cc2)cc1`. The asterisks `*.*.` might be placeholders or errors. I will remove them and try converting the cleaned SMILES string `COc1ccc(Nc2ccc(OC)cc2)cc1`.
Let's start with the cleaned SMILES string of the first reactant.
Action: Smiles to IUPAC
Action Input: c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-22-bromo-9,9-dimethylfluoreneThe IUPAC name for the first reactant is "2-bromo-9,9-dimethylfluorene". Now, I will convert the cleaned SMILES string of the first reactant to its InChI representation.
Action: Smiles to InChI
Action Input: c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2InChI=1S/C15H13Br/c1-15(2)13-6-4-3-5-11(13)12-8-7-10(16)9-14(12)15/h3-9H,1-2H3The InChI representation for the first reactant is "InChI=1S/C15H13Br/c1-15(2)13-6-4-3-5-11(13)12-8-7-10(16)9-14(12)15/h3-9H,1-2H3".
Next, I will convert the cleaned SMILES string of the second reactant to its IUPAC name.
Action: Smiles to IUPAC
Action Input: COc1ccc(Nc2ccc(OC)cc2)cc14-methoxy-N-(4-methoxyphenyl)anilineThe IUPAC name for the second reactant is "4-methoxy-N-(4-methoxyphenyl)aniline". Now, I will convert the cleaned SMILES string of the second reactant to its InChI representation.
Action: Smiles to InChI
Action Input: COc1ccc(Nc2ccc(OC)cc2)cc1InChI=1S/C14H15NO2/c1-16-13-7-3-11(4-8-13)15-12-5-9-14(17-2)10-6-12/h3-10,15H,1-2H3The InChI representation for the second reactant is "InChI=1S/C14H15NO2/c1-16-13-7-3-11(4-8-13)15-12-5-9-14(17-2)10-6-12/h3-10,15H,1-2H3".
I now have all the necessary information for the reactants:
1. First Reactant:
- IUPAC Name: 2-bromo-9,9-dimethylfluorene
- SMILES: c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2
- InChI: InChI=1S/C15H13Br/c1-15(2)13-6-4-3-5-11(13)12-8-7-10(16)9-14(12)15/h3-9H,1-2H3
2. Second Reactant:
- IUPAC Name: 4-methoxy-N-(4-methoxyphenyl)aniline
- SMILES: COc1ccc(Nc2ccc(OC)cc2)cc1
- InChI: InChI=1S/C14H15NO2/c1-16-13-7-3-11(4-8-13)15-12-5-9-14(17-2)10-6-12/h3-10,15H,1-2H3
Final Answer:
1. First Reactant:
- IUPAC Name: 2-bromo-9,9-dimethylfluorene
- SMILES: c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2
- InChI: InChI=1S/C15H13Br/c1-15(2)13-6-4-3-5-11(13)12-8-7-10(16)9-14(12)15/h3-9H,1-2H3
2. Second Reactant:
- IUPAC Name: 4-methoxy-N-(4-methoxyphenyl)aniline
- SMILES: COc1ccc(Nc2ccc(OC)cc2)cc1
- InChI: InChI=1S/C14H15NO2/c1-16-13-7-3-11(4-8-13)15-12-5-9-14(17-2)10-6-12/h3-10,15H,1-2H3
> Finished chain.
{'input': 'I want the IUPAC name, the SMILES and the InChI representations for the reactants of the reaction contained in the image: image.png',
'output': '1. First Reactant:\n - IUPAC Name: 2-bromo-9,9-dimethylfluorene\n - SMILES: c1ccc2c(c1)C(C)(C)c1cc(Br)ccc1-2\n - InChI: InChI=1S/C15H13Br/c1-15(2)13-6-4-3-5-11(13)12-8-7-10(16)9-14(12)15/h3-9H,1-2H3\n\n2. Second Reactant:\n - IUPAC Name: 4-methoxy-N-(4-methoxyphenyl)aniline\n - SMILES: COc1ccc(Nc2ccc(OC)cc2)cc1\n - InChI: InChI=1S/C14H15NO2/c1-16-13-7-3-11(4-8-13)15-12-5-9-14(17-2)10-6-12/h3-10,15H,1-2H3'}
First, it’s worth noting that the model’s reasoning about the tool use is accurate.
The ReAct prompt’s reasoning enables the agent to correctly identify and provide information only about the reactants, contrary to the previous case.
For reactant 2, the extracted information is correct, with both representations that we asked for. On the other hand, for reactant 1, the information provided by the model is not correct at all. But this is because it can not identify the R chain as a proper part of the molecule, something that was partially expected. Thus, the name and representations provided by the agent correspond to a molecule where R corresponds to a hydrogen atom.
In summary, we implemented a basic agent for extracting chemical reactions from images. The agent proved to improve vanilla models by far, and the results can be even better by making more robust tools.
6.1. References#
Madeleine C. Deem, Joshua S. Derasp, Thomas C. Malig, Kea Legard, Curtis P. Berlinguette, and Jason E. Hein. Ring walking as a regioselectivity control element in pd-catalyzed c-n cross-coupling. Nature Communications, May 2022. URL: http://dx.doi.org/10.1038/s41467-022-30255-1, doi:10.1038/s41467-022-30255-1.
Yujie Qian, Jiang Guo, Zhengkai Tu, Connor W. Coley, and Regina Barzilay. Rxnscribe: a sequence generation model for reaction diagram parsing. Journal of Chemical Information and Modeling, 63(13):4030–4041, June 2023. URL: http://dx.doi.org/10.1021/acs.jcim.3c00439, doi:10.1021/acs.jcim.3c00439.